The study involved patients with HR-positive/HER2-negative localised disease, but additional analyses are required to describe implications for clinical practice.
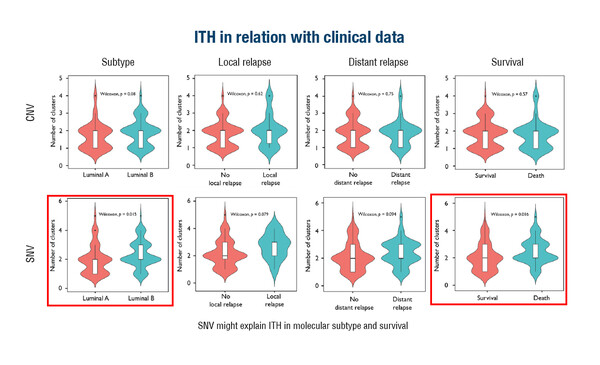
Single-cell DNA sequencing revealed considerable intratumoural heterogeneity in hormone receptor-positive/HER2 receptor-negative localised breast cancer in a study involving 138 patients and presented at the Molecular Analysis for Precision Oncology Congress (MAP) 2023 (Paris, 4–6 October) (Abstract 2O). Most samples had 1–2 clusters of copy number variants (CNV) and 1–3 clusters of single-nucleotide variants (SNV). Local and distant relapse rates were 7% and 28%, respectively, and the survival rate was 64%. There was no relationship between CNV-based clusterisation and luminal subtype or clinical outcomes, but SNV clusterisation was associated with luminal subtype and survival.
In CNV-based clusterisation, specific gene amplifications and deletions were found mainly in tumours with 2 and 3 clusters: deletion events were more prevalent in 3-cluster tumours and in luminal B, compared with luminal A, cancers. Specific genes associated with 2-cluster tumours included AKT1, ATM, KRAS and PIK3CA, while BRCA1, ERBB2, GATA3, IDH1 and NRAS were associated with 3-cluster tumours.
The mutational phylogenies of luminal A and B tumours were different, suggesting that SNV-derived intratumoural heterogeneity may emerge and evolve independently through distinct mutations and clonal patterns.
“These results confirm what has previously been reported with multiregion sequencing (Nature. 2023;616:525–533),” observes Prof. Christos Sotiriou from Jules Bordet Institute, Belgium, discussing the study results. “As expected with a relatively small study, the findings raise questions for further investigation. For example, it is interesting to note the lack of an association between CNV- and SNV-based clusterisation and worse outcomes in terms of local and distant recurrence, which differs from what was found with CNVs and recurrence in the TRACERx non-small cell lung cancer study. Analyses combining somatic mutations and copy number may provide more information. And the group’s ongoing analyses should help to shed further light on the characterisation of molecular differences between luminal A and luminal B subtypes,” he says. Sotiriou concludes: “Looking ahead, to realise its full potential, it is likely that single-cell DNA sequencing will find application in combination with a range of analyses that reflect the incredible complexity of cancer, including RNA sequencing, epigenetics and intratumoural interactions between tumour, stromal and immune cells.”
Abstract discussed:
Tran DT, et al. Dissecting intratumoral heterogeneity in localized HR+/HER2– breast cancer using single-cell DNA sequencing (scDNA-seq). MAP Congress 2023, Abstract 2O
Proffered Paper Session 1, 05.10.2023, h. 13:30 – 15:05 CEST, Auditorium A