Whole-genome sequencing and single-cell sequencing can provide valuable insights not only in the identification of new therapy targets, but also to assess minimal residual disease and overcome treatment resistance
Two presentations at the Molecular Analysis for Precision Oncology Congress (MAP) 2024 (London, 16–18 October) illustrate the relative merits of whole-genome sequencing (WGS) versus single-cell sequencing and provide insights into how these techniques could be used more widely in the treatment of solid tumours. In one study, researchers used data from the UK NHS Genomic Medicine Service’s routine WGS of patients with acute lymphoblastic leukaemia (ALL) and the 100,000 Genomes Project to create a ‘tumour first–germline late’ pipeline (Abstract 93MO). They found that clinically relevant rearrangements in the DUX4 transcription factor – associated with favourable outcomes – were present in at least 5% of all cases. In addition, they were able to identify additional clones, including novel targets, which were not seen with standard next-generation sequencing and may have utility in minimal residual disease monitoring.
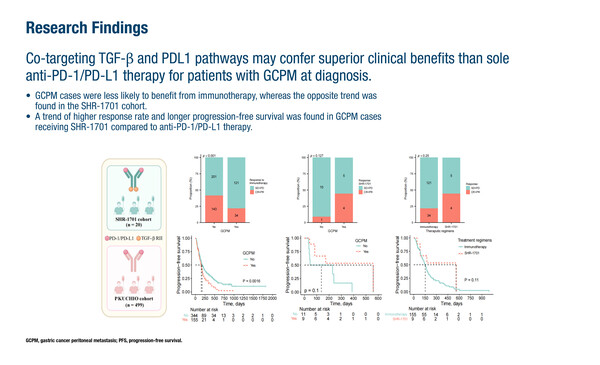
In a second study, single-cell RNA sequencing revealed changes in the peritoneal tumour microenvironment (TME) of patients with gastric cancer and peritoneal metastases (GCPM) before, compared to after, therapy (Abstract 165MO). Matched samples from primary tumours, malignant ascites and peritoneal metastases, along with paired pre- and post-treatment progression ascites samples from 17 patients were subjected to single-cell RNA sequencing and spatial transcriptomics. The study revealed terminally differentiated epithelial MUC1+ cancer cells with a high epithelial-to-mesenchymal transition, which correlated with a poor prognosis. In addition, pro-tumour C1Q+ macrophages were found in the peritoneal microenvironment of ascites samples from patients with treatment-resistant tumours. Crosstalk between C1Q+ macrophages and MUC1+ tumour cells was observed, with enhanced transforming growth factor (TGF)-β production in patients with immunoresistant disease. Applying these findings to a post-hoc analysis of a phase I trial and an immunotherapy cohort indicated that co-targeting TGF-β and PD-L1 pathways with SHR-1701 could be more effective than standard immunotherapy alone.
Prof. Mariam Jamal-Hanjani from the UCL Cancer Institute, London, UK, describes how WGS adds breadth to investigations, while single-cell sequencing provides depth: “WGS is valuable where a comprehensive view of the genome is needed, for example, in cancers known to be associated with rearrangements and copy number alterations, in cancers with germline disposition, for discovery research to identify potential novel drivers and to study cancer evolution in greater detail. In contrast, single-cell sequencing provides more cellular resolution and permits the deeper exploration of aspects such as clonal composition, intratumour heterogeneity and the TME.” The addition of spatial transcriptomics to single-cell sequencing adds even greater depth to the findings by mapping the relationship between the different types of cells within the TME, as well as cell-to-cell interactions, to further inform therapeutic targeting.
For Jamal-Hanjani, the studies presented at MAP 2024 highlight how genomics and transcriptomics can reveal new mechanisms not only for translation into novel therapies, but also to improve minimal residual disease assessment and help circumvent resistance. “Depending on whether breadth or depth is needed, similar studies could be employed in other solid tumour types, particularly those with a poor prognosis where few management strategies currently exist,” she concludes. “However, wider use of WGS and single-cell sequencing in clinical trials and then in practice can only be expanded if current challenges, including high costs and the need for high-quality and fresh tissue samples, can be overcome.”
Programme details
Volkova N, et al. Whole genome sequencing advances aiding clinical care of patients with acute lymphoblastic leukaemia. MAP 2024, Abstract 93MO
Peng H, et al. Single-cell characterization of differentiation trajectories and drug resistance features in gastric cancer with peritoneal metastasis. MAP 2024, Abstract 165MO
Mini Oral Session, 17.10.2024, h. 13:00 – 14:00, Auditorium