As a proof of concept, an AI-powered large language model matching platform shows promise in patient selection
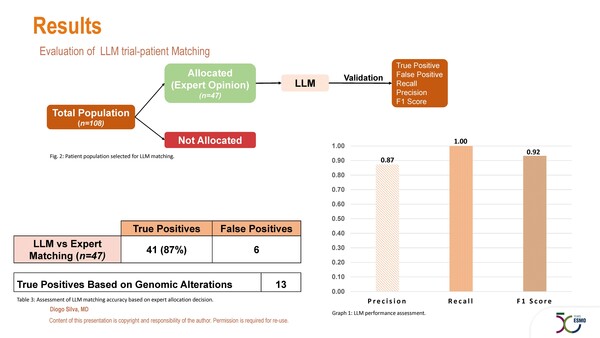
Manual identification of patients for inclusion in clinical trials is laborious and some patients who are eligible can be overlooked in the process. “Here is where artificial intelligence (AI) can play a central role,” observes Prof. Rudolf Fehrmann from the University Medical Center Groningen, Netherlands. “Preliminary data highlight an opportunity for AI to potentially improve patient selection and accrual in trials.” Proof of concept comes from a study presented at the ESMO AI & Digital Oncology Congress 2025 (Berlin, 12–14 November) where an AI-powered platform (MedgicalAI®), leveraging a large language model (LLM), showed that automated eligibility matching of patients referred to a phase I drug development unit is feasible (Abstract 382MO, key results in the box below). In total, 108 patients were assessed and the LLM matching platform identified 41 true positives and 6 false positives, yielding 87% precision for the proportion of AI-generated matches that were truly eligible based on clinical validation. The authors reported high concordance of AI with the allocation decisions of clinical experts, discordances being due mainly to external constraints, such as trial slot availability and incomplete clinical referral data.
“Of course, these findings were observed in a limited set of data, conducted in the context of an experienced phase I clinical trial unit,” notes Fehrmann. “It will be interesting to see how this type of tool would cope with much larger databases involving the complex inclusion and exclusion criteria associated with later-phase trials. In addition, how will it perform outside the specialist setting, for example, in general clinical practice where there may be less comprehensive patient information provided?” Also, the degree of benefit AI can provide over manual selection remains to be assessed. “The study demonstrated high concordance with expert clinician selection but it would be nice to see that it could actually increase and improve patient accrual for studies,” he comments.
AI-based biomarkers have gained prominence in histopathology, where deep learning can identify patterns and transform these into actionable insights for a variety of uses. Also presented at the Congress, a second study highlights the type of technology that could be used to enable researchers to conduct smarter clinical trials, enriching the population with higher-risk patients with early breast cancer (Abstract 1MO, key results in the box below). Data had previously shown that the DeepGrade (DG) tool, based on whole slide histology images and deep learning, could be used to classify Nottingham Histological Grade (NHG)2 samples into DG2-high and DG2-low, where the former had a higher risk of recurrences (Ann Oncol. 2022;33:89–98). In the current study, tile-level DG predictions for 2,522 surgically excised ER-positive/HER2-negative NHG2 breast tumour samples from Swedish centres showed that those with clusters at the tumour front had a better prognosis, with a higher 12-year survival rate than those without clusters at the tumour front (95.2% versus 91.0%; p<0.001). Tumours with front clusters were associated with almost half the risk of progression versus those without front clusters (hazard ratio 0.52; 95% confidence interval 0.32–0.84; p=0.007).
“The field of AI-driven oncology is moving fast and demonstration of its potential is growing. However, as medical oncologists we now need to know that the AI-based tools are reliable and do what they are supposed to do – i.e. stratify accurately,” concludes Fehrmann. “In this regard, it is timely that frameworks, like the ESMO Basic Requirements for AI-based Biomarkers in Oncology (EBAI), are being generated that detail the key criteria needed for appropriate AI-based biomarker validation.”
At a glance:
Silva DJDD, et al. Enhancing phase I clinical trial selection using artificial intelligence: Evaluation of a large language model algorithm in a dedicated drug development unit. ESMO AI & Digital Oncology Congress 2025 - Abstract 382MO
- N=108
- Large language model algorithm:
- Total patients allocated: 47
- True positives: 41
- False positives: 6
- Precision: 87%
- Recall: 100%
- F1 score: 93%
Boissin C, et al. Identification of a very low-risk subgroup in breast cancer using a combination of spatial representation and AI-derived prognostic markers. ESMO AI & Digital Oncology Congress 2025 - Abstract 1MO
- N=2,777 in training set
- 12-year survival: 97.2% for pts without clusters in tumour front vs 93.0% with clusters; p<0.001
- PFS: HR 0.41; 95% CI 0.22–0.75; p=0.004
- N=2,522 in testing set
- 12-year survival: 95.2% for pts without clusters in tumour front vs 91.0% with clusters; p<0.001
- PFS: HR 0.52; 95% CI 0.32–0.84; p=0.007