Presented studies explore different approaches to predict cancer cachexia and inform on the intra-tumour heterogeneity to personalise treatment
There is increasing evidence suggesting that epigenetic changes might represent critical events of cancer initiation (Bioessays. 2025 Mar;47(3):e202400183). Molecular interrogation techniques and machine learning could help harness the wealth of epigenetic information available to enhance diagnosis and patient care in different tumour types, as findings presented at the Molecular Analysis for Precision Oncology (MAP) Congress 2025 (Paris, 15–16 September) show.
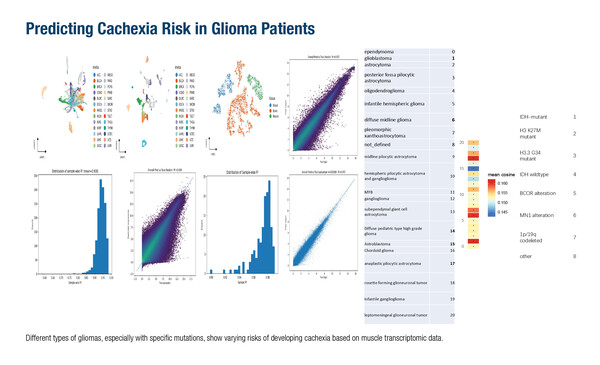
One study developed a novel non-invasive methodology to analyse blood DNA methylation profiles reflecting molecular changes in muscle tissue as a means of predicting cachexia risk in patients with glioma (Abstract 1MO). Around half of all patients with advanced cancer will develop cachexia and regular screening for nutritional risk is recommended, alongside multimodal cachexia care, including nutritional intervention and psychological support (ESMO Open. 2021;6:100092). Recognition of the epigenetic processes that lead to the muscle wasting and metabolic disruption characteristic of cancer cachexia could have a positive impact on patient general wellbeing status.
Using a database of >15,000 samples, a transformer-based deep learning framework was developed and trained to recognise potential signs of cachexia. The model integrated information on CpG site chromosomal positions with functional annotations, and the Genotype Tissue Expression (GTEx) Project database was used to develop a framework to predict gene expression levels across different tissues. This framework was then applied to the blood methylation data from the patient samples to predict virtual muscle transcriptome profiles. The transformer model achieved a high degree of accuracy in predicting gene expression from methylation data (Pearson correlation coefficient 0.89), as well as high specificity for predictions in muscle tissue (Pearson correlation coefficient 0.87). In addition, the authors noted a greater tendency for abnormal expression of cachexia-related genes in predicted muscle transcriptomes from isocitrate dehydrogenase (IDH) wild-type glioma cases than in IDH-mutant glioma cases. This could point to a need to implement more diligent screening and assessment of cachexia as part of routine cancer care in patients known to be at greater risk.
In a second study, analysis of epigenetic changes was explored to unpick the complexity of an individual tumour – high-grade serous ovarian cancer (HGSOC) – marking a step towards personalised therapy for these patients (Abstract 128O).
High-grade serous ovarian cancer (HGSOC) is characterised by a high degree of intra-tumour heterogeneity which, on a molecular level, is linked to progression, treatment resistance and recurrence of HGSOC owing to the co-existence of subpopulations of cells with distinct genetic and epigenetic traits within a single tumour mass (Int J Mol Sci. 2023;24:15077).
The study’s authors used biopsy tissue from patients with HGSOC at diagnosis and again after neoadjuvant chemotherapy and surgery, obtained as part of the prospective SCANDARE biobanking study (NCT03017573) and from other sources. Single-cell RNA-sequencing, single-nucleus RNA-sequencing and single-cell epigenomics were performed with a focus on a specific histone mark, H3K4me1, associated with active or ‘primed’ enhancers and promoters. A total of 13 recurrent tumour transcriptomic phenotypes were identified from >200,000 malignant cells, and longitudinal analyses showed that chemotherapy enriched for cells possessing mesenchymal and inflammatory features and reduced the cells displaying proliferative, hypoxic and interferon-associated states. Using epigenomic data, enhancer ‘priming’ was tracked across these different cell states enabling identification of transcription factors driving resistance to chemotherapy. Accessing the epigenomic landscapes of a wide range of tumour microenvironment populations using single-cell technology enabled the observation of treatment-associated epigenomic changes specifically within a subset of fibroblasts. The study provides proof-of-concept of the use of single-nucleus profiling to offer up valuable information on tumour complexity to inform personalised therapy. A longitudinal study of a larger cohort is underway to further evaluate the predictive value of the tumour state and tumour microenvironment composition at diagnosis.
Programme details
Abstract 1MO - Maimaiti A, et al. DNA methylation-based epigenetic signatures for glioma classification and non-invasive diagnosis: A transformer framework integrating cachexia risk assessment. MAP Congress 2025
Educational session, 15.09.2025, h. 16:15 - 17:45, Auditorium Amphi Lovoisier
Abstract 128O - Landais Y, et al. Evolution of transcriptomic and epigenomic intra-tumor heterogeneity in high-grade serous ovarian cancer with chemotherapy. MAP Congress 2025
Proffered Paper Session, 15.09.2025, h. 14:15 - 15:45, Auditorium Amphi Lovoisier