Results from the TRACERx study suggest a potential role of CHIP as a prognostic biomarker, but further research is needed
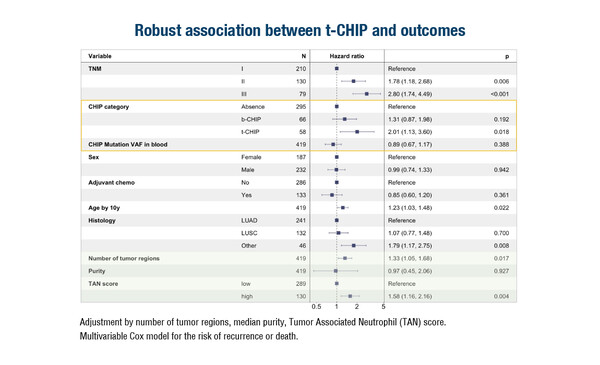
Clonal haematopoiesis of indeterminate potential (CHIP) is associated with unfavourable survival in patients with solid tumours (Cell Stem Cell. 2017;21:374–382). Results from the TRACERx study presented at the Molecular Analysis for Precision Oncology Congress (MAP) 2023 (Paris, 4–6 October) show that CHIP-mutated immune cells infiltrating the tumour microenvironment (TME) (t-CHIP) was an independent predictor of adverse outcomes (hazard ratio 2.0; 95% confidence interval 1.1–3.6) in patients with non-small cell lung cancer (NSCLC) (Abstract 8O).
“This is the first time that CHIP in the TME has been seen to be associated with poor outcomes in lung cancer,” says Prof. Christian Rolfo from the Tisch Cancer Institute, Icahn School of Medicine in New York, USA, and President of the International Society of Liquid Biopsy (ISLB), commenting on the study results.
CHIP was observed in one-third of 421 patients enrolled in the trial and was found across histological subtypes. In a multivariable model, t-CHIP was associated with an increased risk of disease recurrence or death. Among patients with CHIP, 46% had t-CHIP, and this was associated with a skew towards myeloid cells (neutrophils, monocytes).
“These findings open the door for prospective clinical trials looking to confirm the possible role of CHIP as a prognostic biomarker in lung cancer and to monitor disease progression during treatment. The potential for a predictive role for CHIP in different tumour types also warrants study,” says Rolfo.
The study’s methodology comprised extraction of CHIP mutations from whole exome sequencing data of patients’ blood and then genotyping in 1,554 surgical tumour regions. TME cellular compositions were estimated using imaging mass cytometry and RNA sequencing data. Findings were validated on The Cancer Genome Atlas data from patients with NSCLC.
Rolfo concludes, “While incorporating CHIP into liquid biopsies is fairly simple, detection of t-CHIP in tumour samples could be more complicated. It would be useful to investigate the possibility of detecting t-CHIP in liquid biopsies, because this would expedite the process considerably and provide the opportunity to perform these tests in clinical practice.”
Abstract discussed:
Pich O, et al. Clinical implications of clonal hematopoiesis in the tumor microenvironment of non-small cell lung cancer. MAP Congress 2023, Abstract 8O
Proffered Paper Session 2, 05.10.2023, h. 17:00 – 18:20 CEST, Auditorium A