In the MATCH-R study, comprehensive profiling included successful sequencing of DNA and also of RNA, enabling the detection of actionable genomic alterations
Final results from the MATCH-R study presented at the Molecular Analysis for Precision Oncology Congress (MAP) 2023 (Paris, 4–6 October) demonstrated that comprehensive molecular profiling is feasible on a large scale and can help to guide treatment adaptation (Abstract 5O).
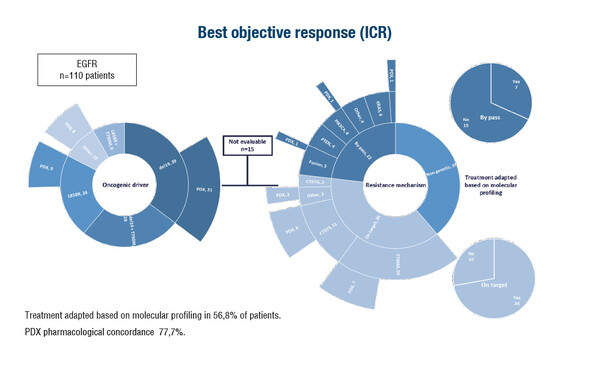
Among 859 patients enrolled in the study (2015–2022), mainly with lung, gastrointestinal and prostate cancers, next-generation sequencing (10< tumour content<30%) or whole exome sequencing and whole transcriptome sequencing (tumour content>30%) was used to investigate 1,144 tissue biopsies before and/or after administration of targeted therapy. The most common molecular drivers were EGFR, FGFR, ALK, BRAF and KRAS. In 66.2% of cases, samples were collected at resistance to targeted treatment, and in 41.0% of them no molecular mechanism was identified and 34.1% showed on-target resistance. The remaining 24.9% exhibited a by-pass resistance mechanism and molecular profiling revealed 60 variants, with PIK3CA (18.3%), KRAS (11.7%), PTEN (11.7%), AKT1 (6.7%), and NRAS (5.0%) being the most altered genes. Treatment was adapted based on molecular profiling in 56.8% of patients with EGFR mutations, 26.0% with FGFR mutations and 56.2% with ALK aberrations.
A total of 282 biopsies were used to establish murine patient-derived xenograft models – by the implantation of tumour fragments into NOD scid gamma mice and amplification up to five passages – with a 40.5% success rate.
“This impressive study in a large cohort – despite being a single-centre study – has important implications in the field of personalised approaches, showing that complete molecular characterisation is feasible in a substantial proportion of patients,” comments Dr Emre Kocakavuk from the University Hospital Essen, Germany. “What is novel at this scale is that the comprehensive profiling included sequencing not only of DNA at the whole exome level, but also of RNA. This is particularly important for the detection of some genomic alterations, such as fusions, that may be missed by other techniques,” he says, suggesting that the future incorporation of molecular signatures may add another useful dimension to this type of profiling.
According to Kocakavuk, the information on treatment adaptations is impressive but as yet incomplete. “To date, much of the data based on molecular profiling has been related to prognostic and predictive utility. In that respect, the data on treatment adaptations reported at this scale is unique. It appears that adaptations are marker dependent, being more likely in patients with EGFR and ALK mutations than FGFR mutations, and it will be interesting to see if this is a function of RNA sequencing. However, the most important information we await is about the clinical outcomes resulting from treatment adaptations based on molecular profiles, which will be required to convince the oncology community of the benefits of this approach.” The study is estimated to complete in 2025.
Abstract discussed:
Beshiri K, et al. Deciphering resistance mechanisms in cancer: Insights from the final report of MATCH-R study with focus on molecular drivers. MAP Congress 2023, Abstract 5O
Proffered Paper Session 1, 05.10.2023, h. 13:30 – 15:05 CEST, Auditorium A