Sequencing data suggest that non-G12C KRAS mutations could mediate intrinsic resistance to treatments in non-small-cell lung cancer, but functional confirmation is required
Acquired resistance to KRAS G12C inhibitors in KRAS-mutated non-small-cell lung cancer (NSCLC) is an emerging issue. Preliminary results of an analysis presented at Molecular Analysis for Precision Oncology Virtual Congress 2021 (MAP 2021 Virtual) proposes one mechanism behind it, providing the first suggestion that intrinsic and extrinsic resistance have the same origin (Abstract 3MO).
G12C is the most common KRAS mutation in NSCLC. The development of KRAS G12C inhibitors over the past 10 years has been hailed as a therapeutic breakthrough and these agents have shown good activity in early-phase clinical trials in lung cancer and in gastrointestinal cancers (N Engl J Med 2020;383:1207–17). The US FDA recently granted accelerated approval for the use of sotorasib in KRAS G12C-mutated NSCLC. However, reports have started to emerge regarding acquired resistance in various solid tumours (N Engl J Med 2021;384:2382–93), which could compromise treatment.
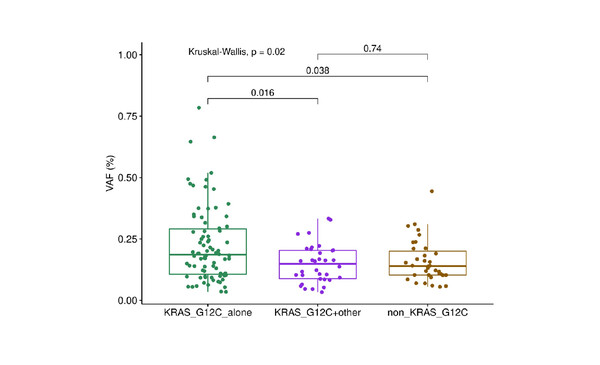
Researchers from La Sapienza University of Rome, Italy, examined multi-sample sequencing data from patients with primary lung adenocarcinomas analysed in the TRACERx study (stage I–IIIA NSCLC) and the international registry AACR Project GENIE. In patients with KRAS G12C mutations, additional KRAS mutations were found in one of seven patients (14%) in TRACERx, and in 42 of 100 patients (42%) in Project GENIE. KRAS variant allele frequency was higher in patients with a KRAS G12C mutation alone, compared with other KRAS mutations and KRAS G12C plus non-KRAS G12C mutations (Kruskal-Wallis p=0.02, post-hoc Dunn test p<0.05 for KRAS G12C mutation alone versus KRAS G12C plus other mutations). There was no difference in tumour mutational burden between patients with KRAS G12C mutation alone and those with multiple oncogenic KRAS mutations (Wilcoxon p=0.7).
“The key implication of these data is that mechanisms that have recently been described for acquired resistance could potentially also be involved in intrinsic resistance,” says Dr Colin Lindsay from The University of Manchester, UK. “Such mechanisms may be present at baseline and are then selected for by treatment, becoming more prominent during the course of therapy,” he explains.
Results may not necessarily be transferrable to advanced cancers, which is the main setting for KRAS G12C inhibitor use at the moment.
However, the sequencing findings presented still need to have functional confirmation, so they should be interpreted with caution. “In patients with KRAS co-mutations identified by sequencing at baseline, we will need to know if there is an increase in variant allele frequency over the course of KRAS G12C inhibitor therapy,” says Dr Lindsay, highlighting the surprising frequency of KRAS co-mutations identified in Project GENIE. “The level (42%) is much higher than we have observed previously, so it would be helpful to understand the reasons for this.” While the study provides some insights into acquired resistance to treatments, according to Dr Lindsay, it is too early to tell how these could affect clinical practice. “This level of sequencing enquiry is still not readily available in the clinical setting. Advanced genomic techniques combined with considerable forward thinking are required to make methods such as single-cell sequencing a scalable reality for most patients,” he says, concluding, “Also, results may not necessarily be transferrable to advanced cancers, which is the main setting for KRAS G12C inhibitor use at the moment.”
Abstract presented:
Marinelli D. Oncogenic non-G12C KRAS mutations in KRAS G12C mutated lung adenocarcinomas in TRACERx and GENIE: A reservoir for intrinsic resistance to KRAS G12C inhibitors? MAP 2021 Virtual, Abstract 3MO
Mini Oral session, 08.10.21, h. 16:00 – 17:00, Channel 1