Molecular pre-screening identified targetable alterations in patients with unknown genomic drivers
“When recruiting for clinical trials, we need to avoid leaving any patients behind – we want to include the right patients for the right targeted treatment,” says Prof. Umberto Malapelle from the University Federico II of Naples, Italy. “This is why molecular pre-screening to include next-generation sequencing (NGS) of circulating tumour DNA (ctDNA) may be a promising approach now under the spotlight.”
Findings demonstrating that this is possible have been presented at the Molecular Analysis for Precision Oncology Virtual Congress 2021 (MAP 2021 Virtual). As shown in an ePoster, researchers from Vall d'Hebron Institute of Oncology, Barcelona, Spain used Guardant360™ to perform NGS of ctDNA to try to identify targetable genomic alterations in 108 patients with a range of solid tumours who were candidates for clinical trials (Abstract 6P). Around one-third had previous tissue NGS and those with a previously known tier I variant according to ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) or known resistance mutations were excluded.
When liquid biopsies are combined together with comprehensive genomic profiling, they provide additional information that can really enhance the clinical picture and therefore optimise decision making.
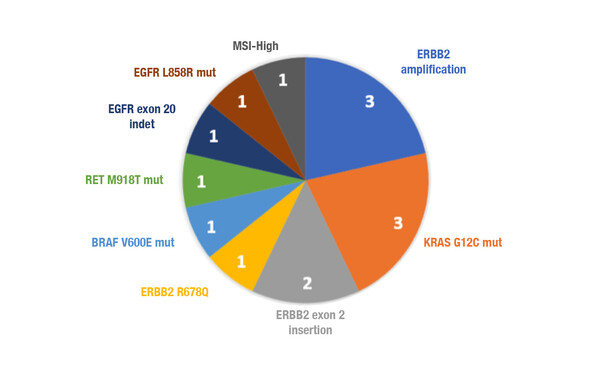
NGS of ctDNA was found to be informative in 89% of the patients tested and the median turnaround time was 8.5 days. Importantly, potentially targetable alterations were identified in 14 patients: 5 had tier I variants and 9 had tier II variants. Five patients received treatment based on the NGS analysis – 4 were treated in a clinical trial and 3 of these patients achieved a partial response. More than 40 tier III variants were identified that could be potential targets in phase I trials. Furthermore, valuable information on RAS-resistant mutations was found in 22 out of 52 patients with colorectal cancer.
“This analysis highlights the further benefits of liquid biopsies,” says Malapelle. “Biopsies from blood and other bodily fluids are not a substitute for tissue-based analysis, but when liquid biopsies are combined together with comprehensive genomic profiling, they provide additional information that can really enhance the clinical picture and therefore optimise decision making.”
Regarding molecular testing, Malapelle highlights how the testing paradigm has now shifted from ‘one gene as one biomarker’ to ‘one mutation as one biomarker’. “Today we have a real-time PCR approach for liquid biopsy analysis, but only by using NGS can we achieve the ability to cover the thousands of alterations to appropriately tailor treatment,” he concludes. “What we should now invest more on is standardisation of protocols from institutions so that we can see more widespread application of NGS of liquid biopsy beyond clinical trials and the research setting.”
Abstract presented:
Alonso G et al. Circulating tumor DNA (ctDNA) next generation sequencing (NGS): Molecular prescreening for tailoring treatment in clinical trials. MAP Virtual 2021, Abstract 6P