Molecular genetic analyses of NETs hint at actionable therapeutic targets and potential predictive biomarkers of response to axitinib
Findings from two studies of neuroendocrine tumours (NETs) presented at the ESMO Sarcoma and Rare Cancers Congress 2023 (Lugano, 20–22 March) help advance the understanding of the molecular landscape of these heterogeneous group of tumours.
The first was a large study developing a multiomics factor analysis (MOFA) model based on combined transcriptome (using master regulator analysis) and methylome data from NETs to identify clinically relevant NET subgroups, potentially paving the way for a molecular classification of NETs and identification of therapeutic targets (Abstract 27O). The transcriptome and methylome data were obtained from RNA and DNA extracted from formalin-fixed, paraffin-embedded (FFPE) tumour samples derived from 194 patients with World Health Organization grade (G) 1–3 NETs. Most biopsies were from the primary tumour (82.5%), and most commonly from the small intestine (27.3%), lung (22.7%) and pancreas (18.0%). MOFA identified 10 factors that were associated with clinical features and patient outcomes. Nine factors could distinguish between the location of the primary tumour site. Factor 6 appeared to differentiate between G1 and G2 tumours of lung origin, as well as being associated with a poor prognosis and patient gender.
“This work is of great value to advance the understanding of NETs as it provides the groundwork to potentially identify NET-specific or subgroup-specific therapeutic vulnerabilities,” explains Dr Melissa Frizziero from Cancer Research UK Manchester Institute. “In contrast to previous studies, which focused largely on selected NET patient sub-populations (e.g. only low-grade NET patient samples or from a specific site of origin), the researchers here assessed a large and comprehensive cohort of tissue samples capturing the whole spectrum of NETs (poorly and well differentiated, from G1 to G3) and different sites of origin, allowing for direct comparisons across subgroups according to histopathological features or locations. In previous studies, transcriptomic and methylation profiling have proven useful to stratify NETs in clinically relevant subgroups (Cancer Discov. 2022;12:692–711; Genome Med. 2022;14:24), holding the potential to succeed where mutation profiling has failed, that is informing personalised treatment for patients with NETs. However, the methodology used herein currently has limited applicability in clinical practice because of the high cost of the molecular biology and bioinformatic approaches applied, and the need for fresh tumour tissue to extract good quality genetic material, which is often difficult to source,” Frizziero highlights.
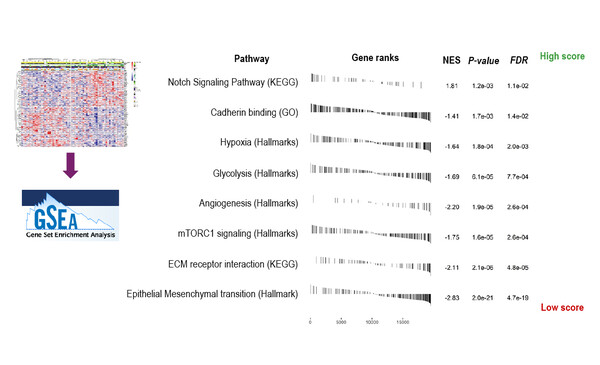
The second study explored transcriptomic signatures to predict response and resistance to the anti-VEGFR inhibitor, axitinib, in patients with advanced extrapancreatic NETs (EP-NETs) enrolled in the AXINET (GETNE 1107) trial (Abstract 28MO). Previously in this trial, axitinib demonstrated a significant improvement in progression-free survival (PFS) in combination with octreotide LAR versus placebo plus octreotide LAR in patients with advanced, progressive G1–2 EP-NETs (Ann Oncol. 2021;32(Suppl5):S906–S920). Using gene expression profiles in 126 FFPE tumour samples from patients recruited in the trial, a gene transcriptomic signature was identified as predictive of PFS with axitinib treatment (p=0.00017). Conversely, the gene signature lacked predictive value in the placebo arm and therefore could be useful to select patients most likely to benefit from this therapy. Underlying molecular pathways potentially involved in response to axitinib were identified as Notch, epithelial-mesenchymal transition (EMT), hypoxia and angiogenesis. Furthermore, dendritic cells, B cells and regulatory T cells were found to be enriched in responders to axitinib treatment, while M1 macrophages were enriched in non-responders.
“Despite demonstrating a PFS benefit for axitinib in patients with EP-NETs, this did not translate into an overall survival benefit compared with somatostatin analogues, suggesting that the efficacy of axitinib might be restricted to a subgroup of patients with EP-NETs,” comments Frizziero. She thinks that this type of analysis can be critical to identify responders to axitinib and can ultimately broaden the treatment armamentarium for a subgroup of patients with EP-NETs. The use of a control arm (placebo plus somatostatin analogue) to support that the predictive ability of the identified gene signature is limited to patients treated with axitinib feeds into the robustness of the approach. However, while the analysis studied potential biomarkers in a large cohort of patients who received treatment with axitinib or placebo, there may be methodological limitations. “RNA extracted from FFPE tumour samples can be of limited quality and the method used for RNA analysis could impact the reliability of the findings,” says Frizziero, adding that multivariable analysis is also needed to identify confounding factors that could affect the results.
While these results are promising, further interrogation into their mechanistic basis is needed, such as an explanation for the involvement of EMT and other signalling pathways in responders to axitinib therapy, and M1 macrophages in non-responders. “The results also need to be corroborated in preclinical models before moving to a clinical prospective validation setting,” concludes Frizziero.
Abstracts discussed:
Carretero-Puche C, et al. Master gene regulation activity and methylation data reveals common patterns in well differentiated neuroendocrine tumors (NETs) from different tumor sites. ESMO Sarcoma and Rare Cancers Congress 2023, Abstract 27O
Proffered Paper Session – Rare Cancers, 20.03.2023, h. 10:50 – 12:10, Hall B1
Lens-Pardo A, et al. Predictive biomarkers of response to axitinib in patients with advanced EP-NETs enrolled in the AXINET trial (GETNE 1107): underlying molecular mechanisms. ESMO Sarcoma and Rare Cancers Congress 2023, Abstract 28MO
Mini Oral Session, 22.03.2023, h. 09:00 – 10:00, Hall B3