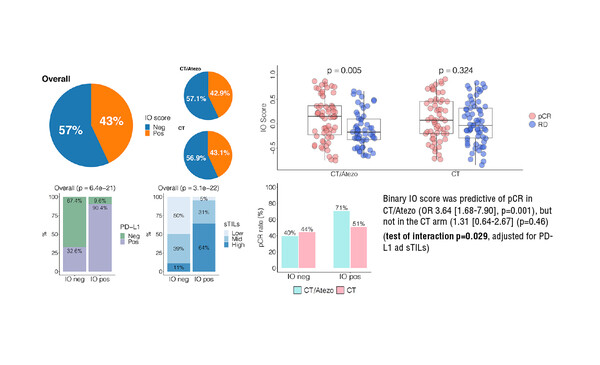
Results from an exploratory post-hoc analysis of the NeoTRIPaPDL1 study show that a 27-gene immune-oncology (IO) score was predictive of the benefit of the atezolizumab combination regimen over chemotherapy alone in patients with triple-negative breast cancer (TNBC) (LBA12). Conversely, the TNBC type did not have predictive value in relation to response rate.
The NeoTRIPaPDL1 study compared eight cycles of nab-paclitaxel/carboplatin with or without atezolizumab in patients with early high-risk, locally advanced triple-negative breast cancer (TNBC). Tissue samples were collected pre-treatment, during treatment (prior to day 1 of cycle 2) and during subsequent surgery, which enabled analysis of potential associations between baseline expression and alterations in tumour and immune features.
Researchers reported an odds ratio (OR) of pathological complete response (pCR) with atezolizumab–chemotherapy of 3.64 (p=0.001) and 1.31 (p=0.46) with chemotherapy alone (interaction p=0.029). Also, there was a significant association (p=0.001) between TNBC subtypes, with the BL1 subtype associated with the highest pCR rate of 70.3% with atezolizumab–chemotherapy and 54.3% with chemotherapy alone, while the LAR subtype was associated with the lowest pCR rates of 22.2% with atezolizumab–chemotherapy and 18.8% with chemotherapy alone (interaction p-value not significant).
Results presented at the ESMO Congress 2021 revealed that a high degree of angiogenesis, elevated fatty acid metabolism and elevated cholesterol homeostasis were independently linked to resistance in the atezolizumab–chemotherapy arm, but not the chemotherapy only arm (interaction p=0.009, p=0.034 and p=0.005, respectively). Glutamine metabolism was linked to resistance only in the atezolizumab–chemotherapy arm. Additionally, super-responders – i.e., patients with no tumour cells detected during treatment – in the atezolizumab–chemotherapy arm tended to have elevated expression of certain immune signatures. Researchers suggested that these patients, accounting for 33.3% of patients in the atezolizumab–chemotherapy arm, could be candidates for de-escalation of therapy.
An elevated expression of a number of immune-related signatures during treatment appeared to be predictive of pCR with either treatment (p<0.01 for both). For example, elevated expression of CD8+ T-cells was associated with respective pCR rates of 58.6% and 61.7% in the atezolizumab–chemotherapy and chemotherapy-only arms, while decreased expression of these T-cells reduced the pCR rates to 22% and 23.1%, respectively.
While immune checkpoint inhibitors, such as atezolizumab, may improve clinical outcomes for patients with a variety of solid tumours, the number of patients who respond is small. These findings showed that combining information from biomarker analysis at baseline and during treatment was more informative than at either timepoint alone, and that this information could help identify patients likely to benefit from immune checkpoint inhibitor therapy.
Bianchini G et al. Predictive value of gene-expression profiles (GEPs) and their dynamics during therapy in the NepTRIPaPDL1 trial. ESMO Congress 2021, Abstract LBA12
Mini oral session – Breast cancer, early stage, 20.9.2021, h. 17:30 – 17:38, Channel 1