Multi-omics approaches incorporating cell-free DNA and circulating tumour DNA show potential in early cancer detection and could help personalise treatment by tumour characteristics
Two studies presented at the ESMO Congress 2021 add to the list of technologies for non-invasive analysis of cancer biomarkers in fluids such as the blood, thus opening new opportunities for the use of liquid biopsy in clinical practice.
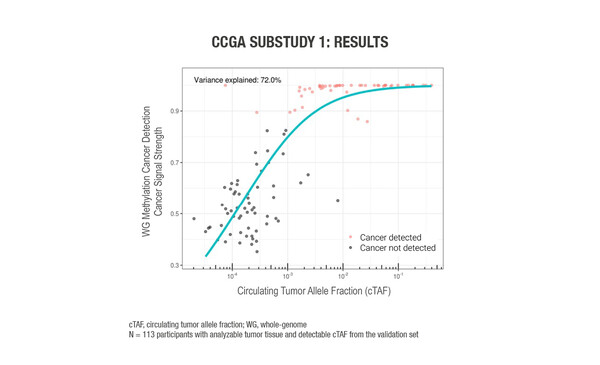
A sub-study of the Circulating Cell-free Genome Atlas (CCGA) study evaluated a range of cell-free DNA (cfDNA) multi-omics as prototype cfDNA-based multi-cancer early detection (MCED) tests using plasma and matched white blood cells and, where available, tumour biopsies (Abstract 1123O). In total, 847 samples were available for analysis – 485 cancer samples and 362 non-cancer samples. Cancer-related features in the form of six cfDNA-omics were used to sample cfDNA: whole-genome (WG) methylation data from WG bisulfite sequencing; single nucleotide variants (SNV) small somatic variant data from error-corrected targeted sequencing in 507 genes; and somatic copy-number aberration (SCNA), fragment length, fragment endpoint, and allelic imbalance data from WG sequencing (WGS). Machine learning was used to ‘train’ the tests to identify a cancer signal via detection of these cancer-related features. A prediction of the origin of cancer signal was also made to inform diagnostic work-up. The WG methylation assay appeared to be the most promising cfDNA approach for a blood-based MCED test, owing to its high (75%) level of accuracy for predicting cancer signal origin and its low signal detection limit that did not require additional sequencing to counterbalance biological background noise from paired white blood cells. Circulating tumour allele fraction (cTAF) accounted for 72% of the variance in WG methylation cancer signal detection feature scores (Figure). In a multivariable analysis, cTAF – but not clinical stage and cancer type – was an independent predictor of cancer signal strength.
Is this approach sensitive enough for all the histologies and sub-groups? The sensitivity is still low in stage I disease in some cancers, and this continues to be a problem.
“This is a very interesting approach in early cancer detection,” remarks Dr Christian Rolfo, from The Tisch Cancer Institute at Mount Sinai Health System, New York, NY, USA. “This study gives us the background to the current US FDA-approved liquid biopsy tool using cfDNA WG methylation presented in the CCGA sub-study and explains the superiority of this method compared with other multi-omics approaches. These data confirm the importance of detecting patients in the early stages of cancer, which will ultimately save more lives. There are still some unresolved questions around this technology. Is this approach sensitive enough for all the histologies and sub-groups? Is it useful to discriminate high-risk populations, such as those with suspected nodules, and cancer? The sensitivity is still low in stage I disease in some cancers, and this continues to be a problem. However, what is true is that this study validates the high specificity of the technique and this is a very important point.” Rolfo thinks that future research efforts should also investigate whether a multi-omic approach beyond cfDNA could identify early-stage cancers and differences between tumour types in order to improve sensitivity.
In a second study – CLIMB360 – prospective and real-time sequencing of circulating tumour DNA (ctDNA) in multiple metastatic cancer types show promise to help identify therapeutic targets in these patients (Abstract 92P). Blood samples from 321 patients with advanced solid tumours – most commonly non-small-cell lung cancer and pancreatic ductal adenocarcinoma – were analysed using next-generation sequencing (NGS). Gene alterations were ranked according to OncoKB therapeutic levels of evidence (LE) from the Memorial Sloan Kettering Cancer Center. Most samples (287 [89%]) had at least one genomic alteration and most were considered pathogenic (268 [83%]). In 26% of samples, the specific pathogenic alteration was associated with LE 1–2 of the same or different histology. “Data from this study characterise molecular aberrations with LE, even though they were created based on tissue NGS, confirming that genomic profiling in different tumour types is feasible and could allow categorisation of patients to receive different targeted therapies. Other tools are already in the clinicians’ hands, such as the ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT), or the MD Anderson criteria, so it will be interesting to know more about how these techniques and tools can be applied together in a prospective study,” says Rolfo.
Despite liquid biopsy being the focus of much research with many techniques undergoing analysis, “Using multi-omics in clinical practice will require a level of expertise for both the techniques themselves and for the bio-informatics analyses that will accompany them. This will have implications in terms of cost, particularly in countries where their use is not reimbursed, thus limiting their accessibility to clinicians and patients,” concludes Rolfo. “Nevertheless, these approaches could change the treatment landscape for some patients, including those with rare tumours by enabling the detection of potentially druggable targets.”
Liu M C et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. ESMO Congress 2021, Abstract 1123O
Proffered Paper session – Translational research, 19.9.2021, h. 13:30 – 13:40, Channel 5
Garcia-Corbacho J et al. First-results of the CLIMB360 study, a prospective molecular screening program across multiple cancer types based on circulating tumor DNA (ctDNA). ESMO Congress 2021, Abstract 92P