Genomic landscaping suggests molecular divergence between cancer subtypes and across disease stages, but molecular similarities between White and Asian populations
Further insights into the molecular landscape of gastric cancer were shared at the ESMO Congress 2021, which could advance the search for novel targets and useful biomarkers for personalised medicine.
Patients with colon cancer had a significantly higher incidence of genomic alterations (median 16%) than pancreatobiliary and duodenal cancers (median 5% for both tumour types), according to data from the GENIE Consortium Cohort, which were used to assess the real-world differential somatic mutational landscape in approximately 12,000 samples from patients with duodenal carcinomas (Abstract 1823P). The frequency of a hypermutant phenotype – defined as tumours harbouring more than 100 mutations – was also significantly higher in colon cancer. Duodenal carcinomas and colon cancer had similar landscapes with only seven differentially mutated genes identified, whereas the landscape between duodenal carcinomas and pancreatobiliary cancers was more divergent with 77 differentially mutated genes identified. These analyses, which scrutinised single-nucleotide variants, small indels, fusions and copy number alterations, as well as gene enrichment and actionability, were performed using the 10th data release of the GENIE Consortium cohort data in 132 duodenal carcinomas, 4,891 pancreatobiliary cancers and 6,968 colon cancers from 19 institutions.
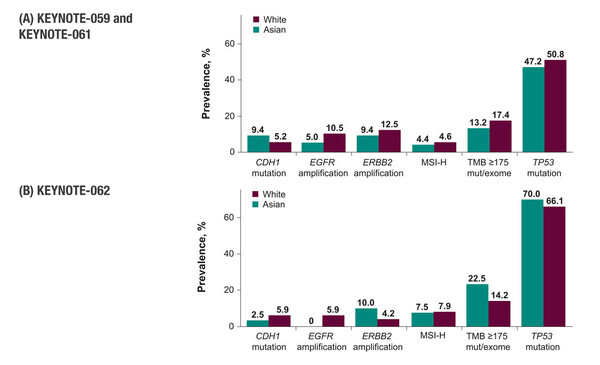
A second genomic analysis explored the landscape of late-stage versus early-stage gastric cancers. Samples from more than 700 patients participating in the phase II KEYNOTE-059 and the phase III KEYNOTE-061 and KEYNOTE-062 studies of pembrolizumab in patients with gastric or gastroesophageal cancers were compared with previously reported subtypes from The Cancer Genome Atlas (TCGA) in early disease (Nature.2014 Sep 11;513(7517):202-9). Alterations in genes of interest – TP53, KRAS, RHOA, CDH1, ARID1A and PIK3CA mutations and EGFR, ERBB2, CD274, CCNE1 and CCND1 amplifications – were evaluated (Abstract 1416P). The prevalence of tumours with chromosomal instability and those classified as being genomically stable was similar in early- and late-stage disease. In contrast, tumours positive for Epstein-Barr virus and those with microsatellite instability were identified at a lower frequency in advanced gastric cancer than in early disease, as reported previously in the TCGA dataset.
Distinct tumour immunity signatures have previously been reported in Asian versus non-Asian patients (Gut. 2015 Nov;64(11):1721-31). In this exploratory analysis, no major differences were reported in the genomic features (Figure) and overall molecular characteristics of Asian and White patients across the subtypes of gastric cancer.
Gastric cancer is a prevalent, heterogeneous cancer with a poor prognosis. In the strive for personalised medicine for this disease, continued molecular characterisation is warranted and future data analyses from these large sample sets are awaited with interest.
Fernandes I. Differential mutational landscape of duodenal carcinomas: a real-world data analysis from the GENIE Consortium Cohort. ESMO Congress 2021, Abstract1823P
Janjigian YY. Genomic landscape of late-stage gastric cancer. ESMO Congress 2021, 1416P