Findings from several studies highlight the increasing utility of this approach to identify druggable genetic alterations
Genetic profiling is becoming an integral part of the diagnosis and treatment of rare tumours. “Important incremental steps have been taken towards a broader implementation of next-generation sequencing (NGS) in this setting to identify druggable alterations, thus ultimately increasing care opportunities for patients with rare malignancies,” confirms Dr Markus Joerger of Kantonsspital St. Gallen, Switzerland, after reviewing some data presented at the ESMO Congress 2021 on the use of molecular profiling in sarcoma, mesothelioma and cancer of unknown primary (CUP).
In one study in patients with sarcoma, DNA- and RNA-based comprehensive genomic profiling was used to analyse the presence of rearrangements (REs), including gene fusions. Several REs were detected; the most common were in EWSR1 (51%), STAT6 (17%), ALK (8%), BCOR (4%) and NTRK1 (4%), with the same RE detected in both DNA and RNA in 66% of samples (Abstract 1532P).
There is still a long way to go but showing that a substantial proportion of patients have targetable genetic alterations is a promising start.
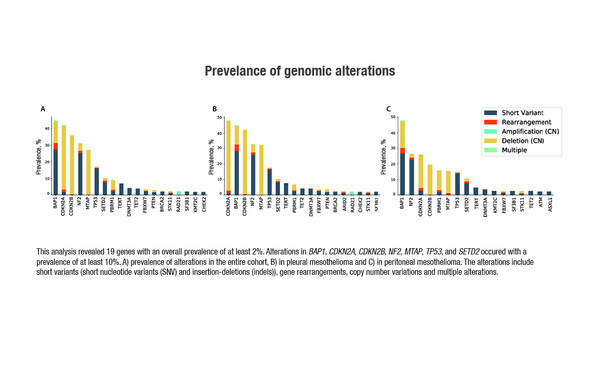
Several gene alterations were also noted in a separate study that utilised NGS to evaluate samples of pleural and peritoneal mesothelioma tumours. In the largest analysis to date in these tumours, genetic alterations with a prevalence of ≥2% of samples were recorded in 19 genes, with at least 10% of patients harbouring alterations in BAP1, CDKN2A, CDKN2B, NF2, MTAP, TP53, and SETD2 (Abstract 1734P).
Taking these results together, Joerger remarks, “The particular challenge with these tumours is that they are inherently difficult to study due to their rarity, and without druggable target alterations, there is limited appeal for conducting adequate clinical trials in this group of patients. By identifying potentially druggable genetic alterations, we are opening the door to specific clinical trials that explore newer targeted anticancer drugs, allowing smaller patient populations and speeding up drug development. There is still a long way to go but showing that a substantial proportion of patients have targetable genetic alterations is a promising start.”
Some data presented also corroborate earlier findings that NGS can be used to biologically characterise CUPs. Whole genome sequencing was used in one study to assess the clinical utility of a CUP prediction algorithm, the performance of which was analysed in a validation cohort of samples with known tumour origin (n=451). The algorithm was applied to 47 patients with diagnostically complex tumours. In 43% (10/23) of patients with CUP and a high-confidence prediction, a probable diagnosis could be reached, suggesting genome-based predictions can be made with a high degree of accuracy (Abstract 1133P). A separate report analysed the mutational profile of patients with CUP who enrolled in the phase II CUPISCO study of targeted therapy/cancer immunotherapy versus platinum-based chemotherapy. Several genetic alterations were identified, including multiple clusters, with at least 30% of samples considered to have potentially druggable genetic alterations (Abstract 1804P). Dr Joerger agrees that these data corroborate earlier findings that NGS can be used to biologically characterise CUP, adding, “Technology has advanced so far that NGS can be implemented in these patients and the resulting data are incredibly valuable for the treating oncologists. Although molecular analysis can suggest the tumour origin, it is not definitive; however, a better understanding of the biology of these cancers is beneficial in making treatment decisions, which is important for these patients.”
Molecular characterisation is increasingly being implemented in the real-world setting. According to Joerger, “The feasibility of hospitals to use the techniques of molecular characterisation or NGS has advanced. The technology has improved enough so that costs and time issues are no longer relevant in many countries and clinical institutions.” However, Joerger concludes, “The complexity of data we are extracting from modern NGS requires adequate expertise within the team treating such patients. Interpretation of genetic data may be assisted by molecular tumour boards or by service packages that provide additional information for the treating clinicians, including the functionality of specific genetic alterations, references to the literature and suggested specific treatments in order to optimise the utility of this approach.”
Ganjoo K et al. Fusion and rearrangement (RE) detection using DNA and RNA-based comprehensive genomic profiling (CGP) of sarcomas. ESMO Congress 2021, Abstract 1532P
Schipper LJ et al. Whole genome sequencing can classify diagnostically challenging tumors. ESMO Congress 2021, Abstract 1133P
Hiltbrunner S et al. Genomic landscape of pleural and peritoneal mesothelioma tumors. ESMO Congress 2021, Abstract 1734P
Westphalen CB et al. Baseline mutational profiles of patients (pts) with carcinoma of unknown primary origin (CUP) enrolled onto CUPISCO. ESMO Congress 2021, Abstract 1804P