Several studies describe pan-cancer approaches utilising the latest technology for better patient selection and outcomes
There is a persistent need for novel, effective biomarkers to predict patients who are most likely to benefit from immunotherapy. The findings from several trials presented at the ESMO Congress 2025 (Berlin, 17–21 October) show that this is currently a field of intense research.
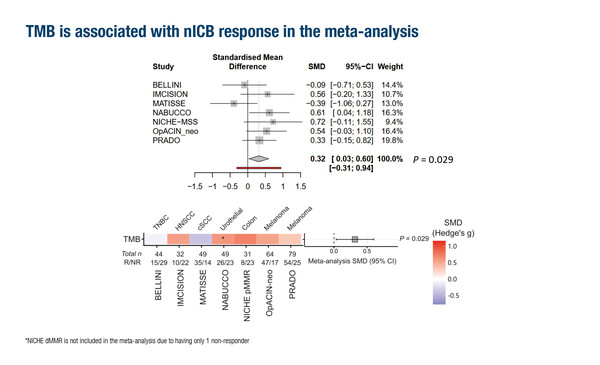
As reported as an oral presentation, a pooled analysis from 7 neoadjuvant immunotherapy trials identified a composite biomarker consisting of tumour mutational burden (TMB), tumour infiltrating lymphocyte score and their interaction (Abstract 112O, key results in the box below). Of 454 patients included in the analysis and treated with either nivolumab plus ipilimumab (90%) or nivolumab alone (10%), response was significantly associated with TMB (p=0.029), higher proliferation (p=0.04) and a number of specific immune-related signatures (p<0.001).
The findings confirm previous work in adjuvant and metastatic settings (reviewed in Cancer Metastasis Rev. 2025;44:58), notes Prof. Olivier Michielin from the Geneva University Hospital, Switzerland. “Interestingly, the analyses presented here are based on whole-exome sequencing and RNA sequencing data,” he says. “While this represents a significant sequencing effort, it involves only next-generation sequencing, a technology to which most centres already have access in large volumes. This improves the likelihood of such agnostic biomarkers being implemented in the clinic to guide patient selection for immunotherapy.”
Deficient mismatch repair (dMMR)/microsatellite instability-high (MSI-H) status was the first tumour agnostic biomarker for immunotherapy, and checkpoint inhibitors are approved for this pan-tumour indication (Cancer Discov. 2017;7:656). Investigation of specific MMR protein loss across 2 large pan-cancer cohorts has provided additional insights to further refine therapeutic approaches (Abstract 111O, key results in the box below). “dMMR tumours are generally rich in neoantigens,” explains Michielin. “Interestingly, this analysis has demonstrated that not all mechanisms leading to dMMR result in the same level of deficiency, so dMMR tumours can be further subcategorised based on the molecular mechanism responsible for their genomic instability. For example, attenuated MSI – often observed with PMS2 and MSH6 single-loss mutations – appears to be associated with poorer benefit of immunotherapy. This finding is likely associated with fewer neoantigens being generated by such tumours,” says Michielin. “If validated, these findings suggest that future treatments could potentially be tailored not only according to dMMR/MSI status, but also to the specific molecular alterations responsible for the instability.”
Among the emerging biomarkers for immunotherapy, human leukocyte antigen (HLA) alterations are increasingly recognised to play a key role in immune escape mechanisms – such as dysfunctional antigen presentation at a tumour surface – that may be responsible for a lack of response to some immunotherapies (J Oncol. 2022;2022:8901326). “If you lose the ability to present an antigen, any attempt to give an immunotherapy targeting that antigen will fail,” explains Michielin. A further study presented at the Congress reported poor outcomes in a cohort of patients with metastatic melanoma and baseline loss of heterozygosity (LOH) of the HLA A*02 allele following treatment with tebentafusp, a first-in-class HLA-restricted therapy targeting this antigen (Abstract 110O, key results in the box below). “The results showed that HLA LOH is not an infrequent finding,” comments Michielin. In the study, 24% of patients from a sample of >50,000 diverse tumour types had somatic LOH of at least 1 HLA allele, and up to 32% of patients with TP53(R175H)-mutant tumours had evidence of LOH of HLA-A*02:01, the most prevalent allele in patients of European ancestry. “Therefore, screening for HLA LOH could potentially be included in future clinical trials or prior to prescribing a therapy that relies on fully functional specific HLA subtypes,” he remarks.
A further consideration in predicting the likely response of a cancer to therapy is an understanding of tumour clonal evolution, as cancer can acquire aggressive traits through a sequential process of genetic alteration, but not all parts of a tumour will evolve at the same rate (Nat Rev Cancer. 2024;24:718–733). “It is extremely complex and resource intensive to sequence every part of a tumour to determine the types of mutation present on a slide,” highlights Michielin. However, computational pathology (CPATH) – a technique involving the digitisation of H&E whole-slide images and use of artificial intelligence to detect clusters of cells with similar histological features – is showing promise as a model to detect genomic changes within a tumour. A study presented in Berlin described the ability of a CPATH model to predict metastatic competence and indolent progression linked to specific genetic alterations in clear-cell renal cell carcinoma (ccRCC) – using a discovery cohort of 1,439 tumour samples from 125 ccRCC specimens, and validation based on 363 ccRCC tumours from The Cancer Genome Atlas (Abstract 109O, key results in the box below). Digital and computational pathology is still in its infancy, and validation of the model is required. “However, a growing body of evidence suggests that slide images can clearly be exploited to a much greater extent than ever thought possible, and with the use of limited resources,” concludes Michielin. “This is groundbreaking research; in future, we could be basing clinical decisions on the findings of computational pathology.”
At a glance:
Tan PB, et al. Pan cancer biomarkers for immunotherapy response: A pooled analysis of neoadjuvant trials. ESMO Congress 2025 - Abstract 112O
- TMB independently associated with response in pan-cancer cohort (p=0.003)
- Significant association between response, higher proliferation (p=0.04) and immune-related signatures, including CXCL9 (p<0.001), Ayers IFN-у (p<0.001) and TIL score (p<0.001)
- Composite response biomarker: TMB (p<0.003) + TIL score (p<0.001) + their interaction (p=0.008)
Gormally MV, et al. Pan-cancer mapping of HLA class I loss of heterozygosity in a 50,000 patient cohort reveals prognostic clinical implications for HLA-restricted therapies. ESMO Congress 2025 - Abstract 110O
- LOH of ≥1 HLA allele: 24% of tumours
- LOH of HLA-A*02:01: ~32% of TP53 (R175H) mutant tumours
- Baseline LOH (A*02 allele) vs no LOH: shorter
- PFS (median 1.2 vs 5.5 months; HR 6.3; log-rank p=1.9e-4) and
- OS (median 5.4 vs 19.5 months; HR 7.2; log-rank p=2.2e-4)
Randrian V, et al. The mechanism of mismatch repair deficiency (MMRd) informs survival outcomes derived from immune checkpoint blockade (ICB) across MMRd solid tumors. ESMO Congress 2025 - Abstract 111O
- Discovery (MSK) cohort (N=1,958)
- MLH1/PMS2 loss: 67%, MSH2/MSH6 loss: 22%, MSH6 single loss: 6%, PMS2 single loss: 5%
- PMS2 and MLH1-epigenetic losses had worse OS vs MLH1-genetic loss (HR 3.5 [1.7–7.0] and HR 2.0 [1.2–3.4], respectively; p=0.0011)
- Validation (CARIS) cohort (N=13,421)
- MLH1/PMS2 loss: 75%, MSH2/MSH6 loss: 11%, MSH6 single loss: 5%, PMS2 single loss: 7%
- Multivariate analysis in pts with MMRd cancers, strongest prognostic factors under ICB therapy were
- Longer mOS: MSI-H, Lynch status
- Shorter mOS: MSS, cancer type (non-GI, genitourinary and rare cancer types)
Spencer C, et al. Capturing clonal evolution through histology. ESMO Congress 2025 - Abstract 109O
- Two-step stratification workflow for adjuvant therapy:
- Predict 9p (AUROC 0.91) or 14q loss (AUROC 0.93), to determine metastatic risk
- Predict PBRM1 mt in all 9p wild-type regions to determine progression risk